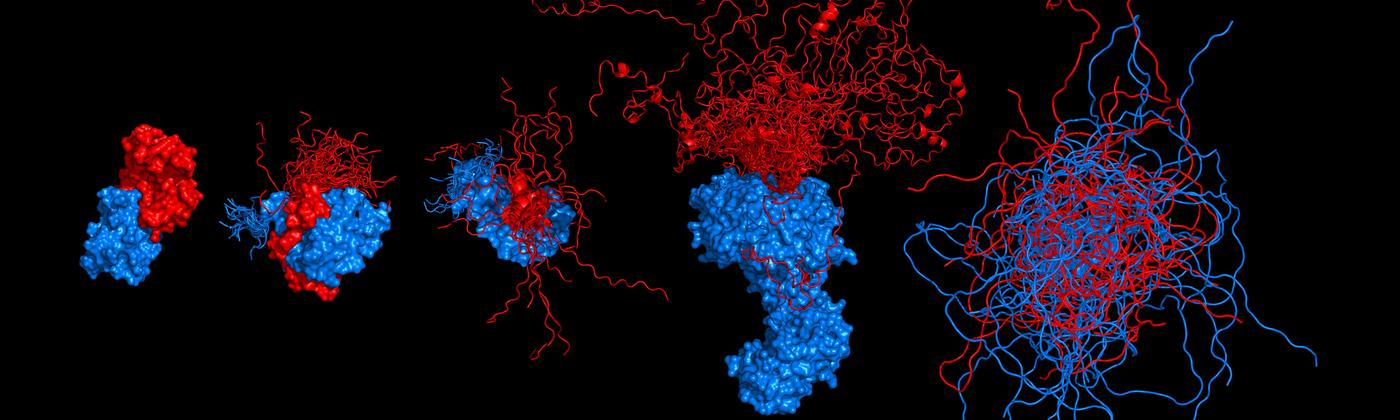
Protein dynamics and intrinsically disordered proteins
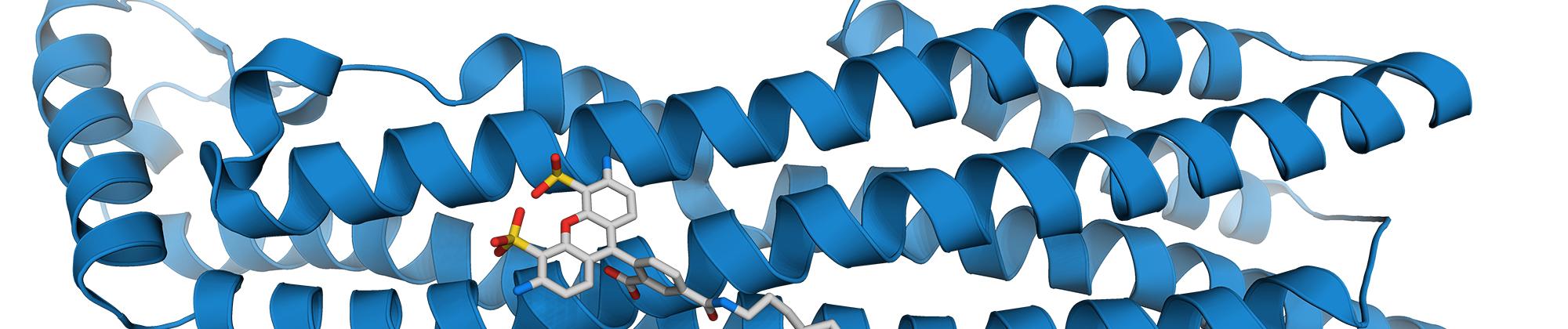
We study fundamental aspects that govern protein structure, dynamics, and function in vitro and in vivo. Towards this goal, we integrate the information on molecular distances, forces, and dynamics from single-molecule spectroscopy with other methods, frequently in close combination with theory and simulations. These approaches have allowed us to map intramolecular distance distributions, determine nanosecond dynamics that govern the diffusive search of protein on their free energy surfaces, and investigate folding and misfolding over a wide range of timescales. Single-molecule methods are also ideally suited for probing the structure, dynamics, and functions of intrinsically disordered proteins. Key methods we employ are single-molecule Förster resonance energy transfer (FRET), photoinduced electron transfer (PET), and optical tweezers.
Effects of the cellular machinery on biomolecular structure, dynamics, and interactions
Many aspects of the physical principles governing protein folding and biomolecular interactions in vitro have been elucidated in the past decades. However, our mechanistic understanding of how cellular components affect the underlying free-energy landscapes has remained limited, largely due to a lack of suitable methods. We investigate the role of cellular factors on protein structure, dynamics, and interactions with single-molecule spectroscopy, ranging from macromolecular crowding and complex reconstituted systems to measurements within live cells. A detailed investigation of these effects will be crucial for understanding the fine balance that governs protein folding and misfolding, the interactions of intrinsically disordered proteins, and the large number of diseases associated with these processes.
Single-molecule method development
A wide range of single-molecule instrumentation is available in the group, including several state-of-the-art confocal instruments with picosecond single-photon counting electronics and a variety of laser sources, a TIRF microscope, and quadruple-trap optical tweezers. However, many projects require the development of novel instrumentation and data analysis tools. Extending the single-molecule toolbox and the accessible timescales is thus a continuous effort, and examples of our developments include nanosecond fluorescence correlation spectroscopy for the measurement of very fast dynamics; the design of microfluidic mixing devices for nonequilibrium single-molecule experiments; novel analysis methods for extracting dynamics from photon statistics; or single-molecule measurements in live eukaryotic cells.
Protein biochemistry
A key requirement for single-molecule experiments is the availability of biomolecular samples of high quality that enable us to probe and specifically manipulate the systems of interest. We use modern molecular biology methods, recombinant heterologous expression, advanced protein biochemistry, and high-resolution purification and chromatography techniques to generate the protein and nucleic acid samples of choice. We also develop and adapt methods to improve specificity and versatility of biomolecular conjugation, such as the use of non-natural amino acids for fluorophore incorporation. Being able to directly combine advanced single-molecule techniques with versatile sample preparation is an important strength of the research in our team.