Pictures

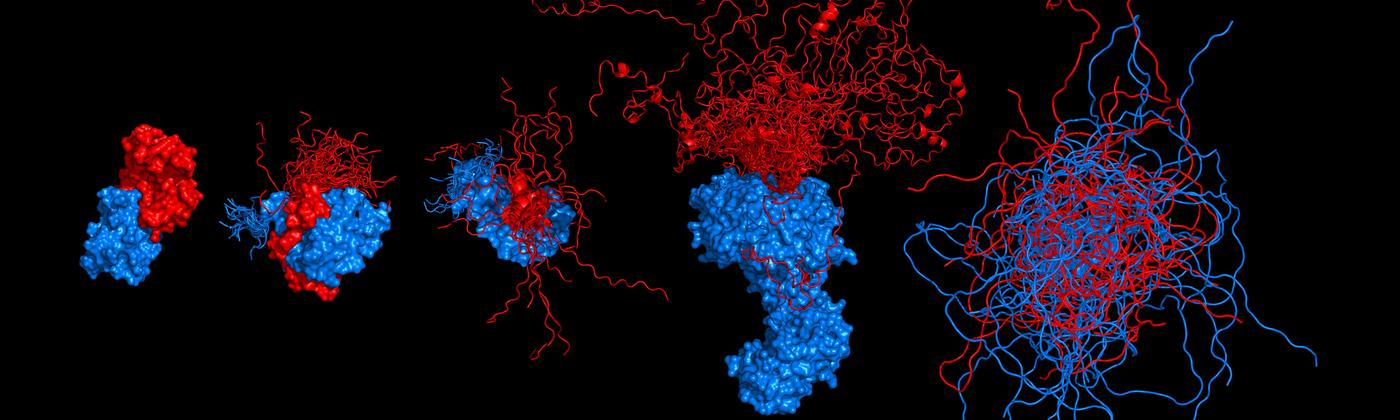
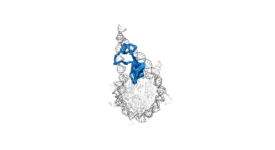
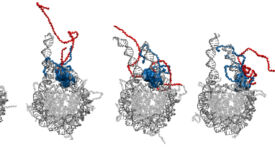
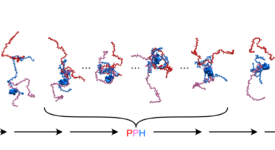
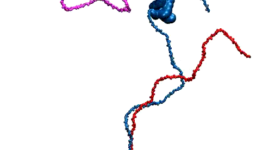
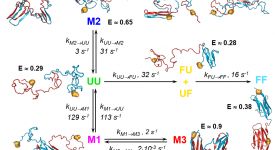
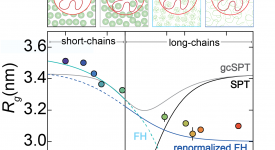
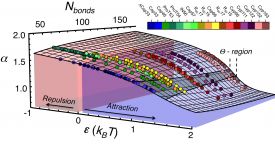
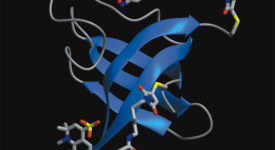
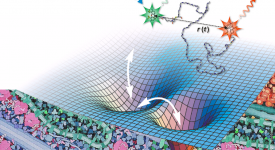
Competitive substitution via ternary complex formation of H1 and ProTa without exchange (Sottini et al. 2020)Competitive substitution via ternary complex formation of H1 and ProTa without exchange (Sottini et al. 2020)
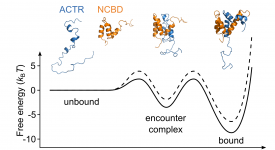
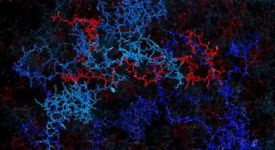
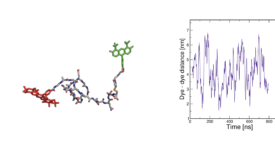
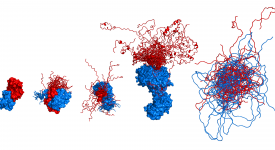
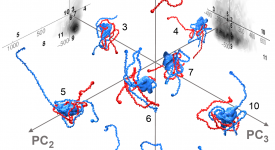
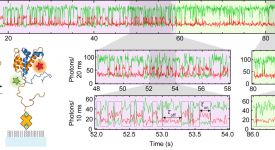
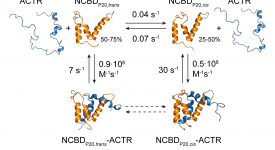
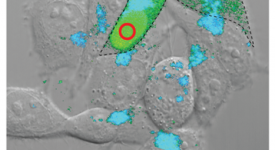
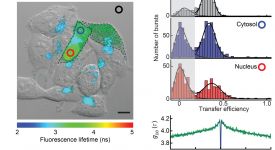
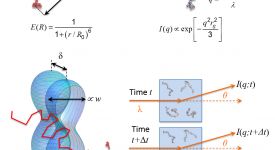
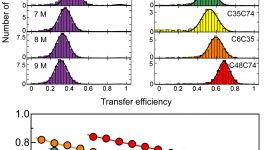
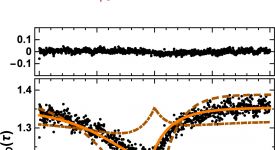
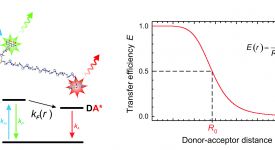
Transition path time measurements reveal an encounter complex in coupled forlding and binding (Sturzenegger et al., 2018)
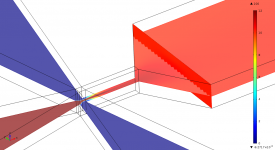
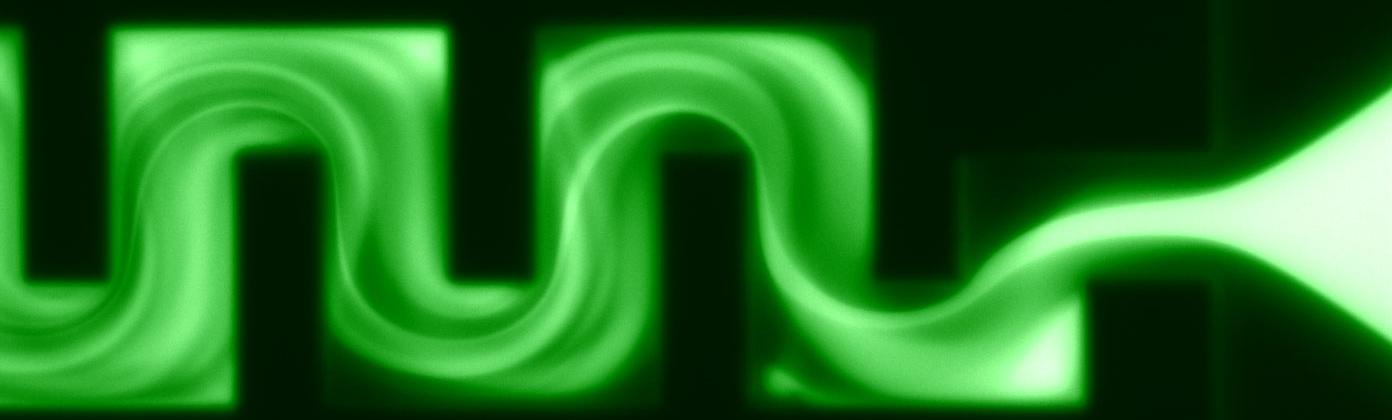
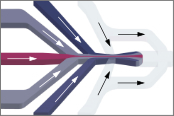
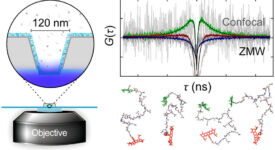
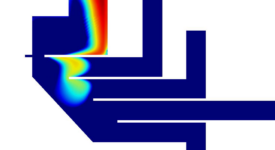
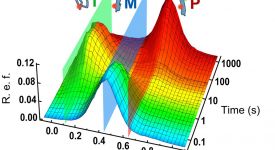
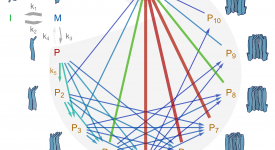
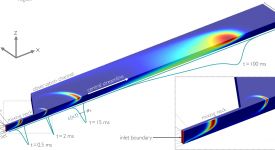
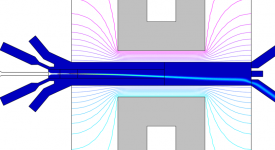
Finite-element calculations of microfluidic mixing by hydrodynamic focusing (Wunderlich et al. 2013)